Corpus Journal of Dairy and Veterinary Science
[ ISSN : 2833-0536 ]
The Microbiology of Cheese and Dairy Products is a Critical Step in Ensuring Health, Quality and Typicity
1Stazione Zoologica Anton Dohrn, Fano Marine Centre, Viale Adriatico, 1-N, 61032 Fano, Italy
2Department of Environmental Sciences, Università di Siena, Via Mattioli, 4, 53100 Siena, Italy
Corresponding Authors
Keywords
Abstract
Cheese and dairy products require a rigorous observation of the procedures, strictly linked to the different indigenous components, such as the initial raw materials, the process chain, the ripening temperature, the water activity (aw), the pH and the contamination of the environment and operators. Microorganisms are key agents in the transformation of milk and in the subsequent phases which confer typicity and stability to cheese and dairy products. Contamination by pathogenic microorganisms may occur, compromising the safety of the final products. Meanwhile, beneficial microorganisms present in cheese and dairy products can produce antimicrobial compounds, thus avoiding spoilage of the products, ensuring their safety for human consumption. This mini review reports a description of the microorganisms involved in the fermentation of milk and in the subsequent processes concerning cheeses and derivatives, highlighting the aspects that microorganisms play in terms of quality, typicality and safety. New aspects emerged from this study, suggesting possible insights and future research. These include cultural approaches on the one hand, which allow for the isolation and characterization of new microbial strains that confer peculiarities in terms of quality and typicality to cheeses and dairy products, and the isolation of lactic acid bacteria that produce bacteriocins as important tools for combating microbial pathogens. On the other hand, investigations on cheese and dairy products using metagenomic approaches with DNA extraction followed by amplification and sequencing of microbial genes, allow the description and monitoring of the entire microbiota involved in the transformation processes of cheese and dairy products. Therefore, the combination of cultural dairy microbiology and metagenomic approaches can lead to improving the characteristics of cheeses and dairy products, while maintaining respect for traditions.
Introduction
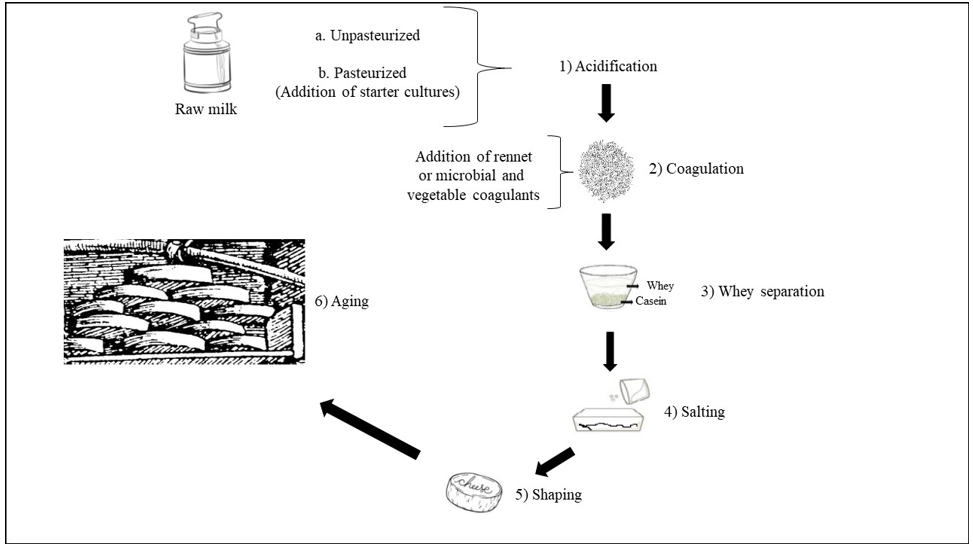
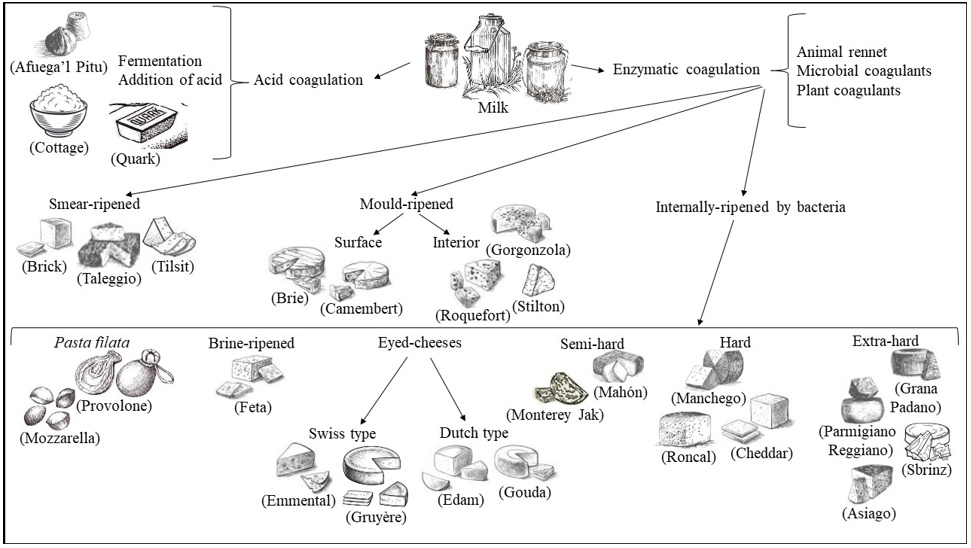
Dairy products include milk-based food, such as cheese, butter, yogurt, kefir, ice cream, and condensed and powdered milk. Among dairy products, cheese is the most varied and widespread. Dairy products present differencies in terms of raw materials, process chain, ripening temperature, water activity (aw), pH, environment and contamination by operators, and all these aspects interact closely with microbial communities of dairy products [1]. Various microorganisms from the environment, including bacteria, yeasts, molds, viruses and bacteriophages, can enter the dairy supply chain in several stages and can affect the entire dairy supply chain [1]. Microorganisms are the key agents in the critical stages of optimizing the overall quality and safety, flavor, appearance and typicality of cheese and dairy products. The term cheese describes dairy products with the definition of “a ripened or unripened, soft, semi‐hard, hard, or extra‐hard, dehydrated milk‐derived product in which the whey protein/ casein ratio does not exceed that of milk” [2]. Cheeses comprehend a great variety of forms, sizes, textures, aromas, and tastes. Production of cheese follows general procedures that include milk acidification; milk coagulation by adding different agents, to be chosen from animal rennet, microbial or vegetable coagulants; cutting the curdled milk originated by coagulation into small pieces; whey drainage; washing; heating to temperatures between 30 °C and 55 °C; pressing and adding salt at concentrations between 1% and 5% NaCl; shaping; aging with maturation and dehydration of the product (Figure 1). Treatments with natural compounds such as immersion in oil, wine or brine, or the addition of ash, flour, spices or vegetable dyes to the packaging of the final product, can also be included [3-5]. When milk is subjected to an acidic coagulation via fermentation of lactic acid bacteria colonizing milk or via addition of acid, it is obtained the production of soft cheeses as Cottage, Quark, Queso Blanco and Afuega’l Pitu (Figure 2).
In the case that milk achieves coagulation via enzymatic treatment, by adding animal rennet or microbial coagulants or plant coagulants to pasteurized or unpasteurized milk, different cheeses can be obtained via three distinct processes: smear-ripening, mould[1]ripening and internally-ripening by bacteria. Smear-ripened cheeses develop a viscous, red-orange smear on their surfaces during ripening. For this reason, they are also called red-smear cheeses or bacterial surface-ripened cheeses. Typical smear-ripened cheeses include Tilsit, Limburger, Romadour, Chaumes, Taleggio, Brick (Figure 2). Mould[1]ripened cheeses are divided in surface mould-ripened, soft and creamy with a white skin cheeses including Brie, Camembert and Carré de l’Est; or in interior mould-ripened, as in the case of Roquefort, a classic blue cheese made from ewe’s milk, and other blue-veined cheeses as Stilton, Cabrales and Gorgonzola (Figure 2). Cheeses internally-ripened by bacteria comprehend many different kinds, evidencing diverse aging and compactness. Pasta filata, ‘spun paste’, cheeses undergoing a single processing with the spinning of the curd dipped in hot water or brine until a semi-flowing plastic consistency is obtained, as in the case of Mozzarella, Provolone, Scamorza and Caciocavallo, mainly from Southern Italy. Brine-ripened cheese refers to cheese that is matured in brine, giving the cheese good stability and inhibiting growth of undesired bacteria, even in warmer climates. Brine-ripened cheeses are mainly produced in countries bordering the Mediterranean Sea and in the Balkan countries and comprehend Beyaz peynir, Sirene, Domiati, Feta and Halloumi cheeses. Eyed-cheeses can be hard-cooked Swiss-type cheese such as Emmental and Gruyere, heating to around 52 °C to remove moisture and harden the curd or Dutch-type semi-cooked cheeses such as Gouda, Edam and Maasdam. Semi[1]hard cheeses comprehending Mahón from the island of Menorca (Spain), Monterey Jack, from California (USA), semi-hard cheese and Caerphilly, a crumbly white cheese originating from the town of Caerphilly, Wales (UK), with all three of these cheeses from cow’s milk. Hard cheeses, with Manchego originating from milk of the sheeps in the La Mancha region of central Spain; Roncal, a Spanish cheese from the Roncal Valley, north Navarra in the Basque territory near the border of France; Cheddar, with its origin in Leicestershire county, in the East Midlands (UK); Ras cheese, the main traditional hard cheese in Egypt. Extra-hard cheeses include Grana Padano, originating in the Po river Valley in northern Italy; Parmigiano Reggiano with a defined geographical area comprehending the territories of the Provinces of Bologna to the left of the Reno River, Mantua to the right of the River Po, Modena, Parma and Reggio in the Emilia Region (Italy); Sbrinz, produced primarily in the Central Swiss cantons of Obwalden, Nidwalden, Lucerne and Zug (Switzerland); Asiago, produced around the alpine area of the Asiago plateau in the regions of Veneto and Trentino-Alto Adige (Italy) (Figure 2).
Microorganisms in Cheese and Dairy Products Transformation
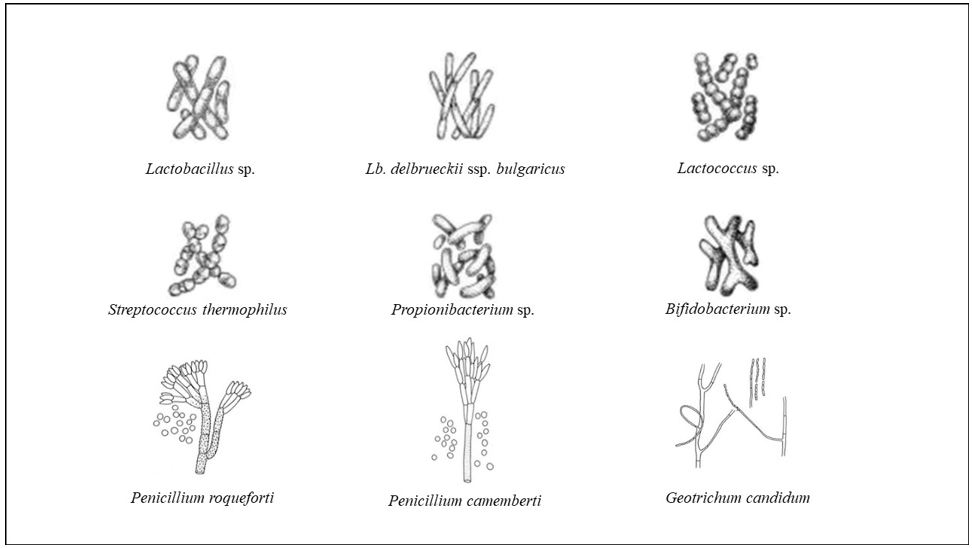
Microorganisms are responsible for the fermentation of milk and the numerous biochemical reactions that occur during production and maturation, which give rise to the characteristic textures and flavors of the cheese [5]. Microorganisms play essential roles in the transformation of milk, in the production and maturation of cheeses, making an important contribution to the development of organoleptic properties through their metabolism and various enzymatic activities. Microorganisms also contribute to the microbiological safety of cheese through the barrier effects of complex microflora and the production of numerous low molecular weight antimicrobial compounds [6]. From the microbiological point of view, cheese and dairy products, both fermented in a natural way and with the addition of starters and/or adjunct cultures, contain a complex mixture of microbial communities, including those relevant for carrying out the processes, or responsible of cheese deterioration, opportunistic and pathogenic organisms, which develop and change during production and maturation [7,8]. From the systematic point of view, cheese bacteria as lactic acid bacteria, enterococci and staphylococci belong to the phylum Firmicutes; bifidobacteria, propionibacteria and corynebacteria to the phylum Actinobacteria; enterobacteria to the phylum Proteobacteria [9,10]. Archaea have rarely been found in the cheese microbiota, with few members belonging to the genera Thermocladium and Sulfurisphaera of the phylum Crenarchaeota, and to the genus Methanohalobium of the phylum Euryarchaeota [11,12]. Within eukaryotes, yeasts are present in cheese with the genera Geotrichum, Debaryomyces, Kluyveromyces, Candida, Yarrowia, filamentous fungi in cheese include members of the species Penicillium camemberti, P. roqueforti and others Penicillium spp., which are abundant in mould‐ripened cheese varieties [13-15]. The species of filamentous fungi Fusarium domesticum, Scopulariopsis flava and Sc. casei were also detected, even if in low quantities, on the surface of most cheeses [13,16,17]. Except P. roqueforti, all these mold species are highlighted only in the cheese, suggesting a possible domestication of fungi to this particular habitat. P. camemberti likely derives from the wild ancestor Penicillium commune through adaptation processes, as reduction of reproductivity, reduction of mycotoxins production, reduction of pigmentation and a shift from earthy to cheesy in the volatile compounds production [18]. Among bacteria, the phenomenon of adaptation via domestication was detected in Lc. lactis ssp. lactis and in Lc. lactis ssp. cremoris strains [19,20]. These, and the domesticated strains of other lactic acid bacteria species found only in milk and dairy products, seem to have emerged recently due to the selective pressure imposed by the dairy technologies. Over the centuries, strains of lactic acid bacteria, yeasts, and molds have evolved to their domesticated roles, leading to genome decay, loss of pathways, acquisition of genomic elements, and beneficial mutations that provide an advantage in their nutrient-rich food environments [21]. In Figure 3, some morphologies of microorganisms present in cheese have been reported.
Lactic Acid Bacteria
Lactic acid bacteria are the most important microorganisms in milk fermentation, converting lactose to lactic acid, which results in an increased acidity that makes growth conditions of microorganisms other than lactic acid bacteria increasingly unfavourable. Lactic acid bacteria are Gram-positive bacteria with a low Guanine+Cytosine (G+C) content in their DNA. They are acid tolerant bacteria, non-motile, non-spore forming and have rod- or cocci-shaped morphologies [22]. The most common lactic acid bacteria present in milk include the genera Lactobacillus, Lactococcus, Leuconostoc, Streptococcus, Pediococcus and Weisella [23,24]. Lactic acid bacteria need carbohydrates to produce the energy necessary for growth. In milk and dairy products, lactose is the main natural carbohydrate. Lactose is made up of glucose and galactose. The fermentation of lactose is called glycolysis or glycolytic pathway. Obligatory homo-fermentative lactic acid bacteria are able to ferment lactose into pyruvic acid, which is then reduced to lactic acid by the reducing power previously produced in the form of NADH. Thus, in obligatory homo-fermentative lactic acid bacteria, lactic acid is obtained as the sole product, with a chemical pathway with 1 mole of glucose giving 2 lactic acid moles and 2 ATP moles, via this process called homo-lactic fermentation [25]. Obligatory homo-fermentative lactic acid bacteria include strains of the species Lactobacillus acidophilus, Lb. amylophilus, Lb. bulgaricus and Lb. helveticus [25]. Homo-lactic fermentation should theoretically produce 2 moles of lactic acid per mole of glucose consumed with a theoretical yield of 1 g of product per g of substrate. In reality, a part of the carbon source is used by the bacteria themselves for the production of biomass and therefore the experimental yields are generally lower [25]. Hetero-lactic fermentation is the process that is characterized by the formation of co-products such as CO2 , ethanol and/or acetic acid in addition to lactic acid as the final product during the fermentation-phosphoketolase pathway. The first step of glucose degradation, which is called pentose phosphate pathway, leads to glyceraldehyde 3-phosphate, acetyl-phosphate and CO2 . Glyceraldehyde 3-phosphate enters the glycolysis through which it is transformed into lactic acid, while acetyl-phosphate is converted into acetic acid and/or ethanol. In the hetero-fermentative pathway, glucose can give lactic acid and CO2 and ethanol and ATP; or glucose can give lactic acid and CO2 and acetic acid and 2 ATP and 2 NADH. Microorganisms that use only this metabolic pathway for the consumption of carbohydrates are called obligatory hetero-fermentative, among which are Lactobacillus brevis, Lb. fermentum and Lb. reuteri [26,27].
generally lower [25]. Hetero-lactic fermentation is the process that is characterized by the formation of co-products such as CO2 , ethanol and/or acetic acid in addition to lactic acid as the final product during the fermentation-phosphoketolase pathway. The first step of glucose degradation, which is called pentose phosphate pathway, leads to glyceraldehyde 3-phosphate, acetyl-phosphate and CO2 . Glyceraldehyde 3-phosphate enters the glycolysis through which it is transformed into lactic acid, while acetyl-phosphate is converted into acetic acid and/or ethanol. In the hetero-fermentative pathway, glucose can give lactic acid and CO2 and ethanol and ATP; or glucose can give lactic acid and CO2 and acetic acid and 2 ATP and 2 NADH. Microorganisms that use only this metabolic pathway for the consumption of carbohydrates are called obligatory hetero-fermentative, among which are Lactobacillus brevis, Lb. fermentum and Lb. reuteri [26,27].
From a metabolic point of view, lactic bacteria are fastidious microorganisms and are unable to synthesize many amino acids, vitamins and nucleic acid bases. Lactic acid bacteria require 6 to 14 different amino acids, depending on which species a strain belongs to. Free amino acids in milk are limited and amino acids are present as protein components, so the growth of lactic bacteria requires hydrolysis of milk proteins [33]. The hydrolysis of peptides into free amino acids and the subsequent utilization of these amino acids is a central metabolic activity in lactic acid bacteria. Proteolysis by lactic bacteria is therefore the key process influencing the rate of flavor and texture development in most cheese varieties. The breakdown of milk proteins into peptides is catalyzed by proteolytic enzymes present in lactic acid bacteria and the peptides are then further hydrolyzed by exopeptidases and endopeptidases into small peptides and amino acids [34]. In milk, the breakdown of fat releases free fatty acids and glycerol, monoacylglycerides or diacylglycerides. Some free fatty acids are peculiar aromatic compounds in some cheeses, such as in the case of goat cheeses. Free fatty acids can react with free alcohols or sulfhydryl groups to form esters and thioesters, respectively, or act as precursors to numerous other beneficial compounds, such as lactones [35]. Esterase activity was found in various lactobacilli [36], and this is an important feature as the esters contribute to the characteristic flavor of the Swiss type [37] and White-brined cheese [38].
Starter and Non-Starter Microbial Cultures
Microorganisms can grow in milk left at room temperature with a prevalence of mesophilic lactococci [7]. These microorganisms can be maintained by inoculating fresh milk with part of the previous day’s fermented product, and this was at the basis for the production of fermented dairy products for centuries, even before it was known that the bacteria were actually involved. The isolation of lactic acid bacteria from the milk environment began in the second half of the nineteenth century, with the development of starter cultures for the production of fermented dairy products [39]. Starter cultures may contain mixed strain cultures with unknown strains or can be composed of pure and well characterized strain cultures from which isolated bacterial strains can be obtained and maintained at -80 °C or lyophilized for successive utilization as starter cultures [40]. Starter cultures drive the production of acid during the beginning of the fermenting process, nevertheless, these same bacteria may be involved in cheese ripening by producing enzymes involved in proteolysis and the conversion of amino acids into aromatic compounds [41]. Starter bacteria are able to acidify milk producing sufficient acid and reducing the pH to values <5.3 in about six hours and at temperatures in a range from 30 to 37 °C. Depending on the cheese to be prepared, mesophilic or thermophilic starter cultures can be used. The production of Cheddar, Gouda, Edam, Blue and Camembert, requests the addition of mesophilic cultures, whereas cooked hard cheeses such as Emmental, Gruyere, Parmigiano Reggiano and Grana Padano require the milk to be heated to around 50-55 °C and therefore demand the addition of thermophilic starter cultures. The most frequent used starter bacteria originated from one or more of the species Lactococcus lactis, Streptococcus thermophilus, Lactobacillus helveticus and Lb. delbrueckii and the choice of the starter culture depends on the type of cheese to be produced. As an example, the mixed-strain starters for the manufacture for Gouda cheese are composed of acid-forming lactococci, Lactococcus lactis subsp. lactis and L. lactis subsp. cremoris, together with citrate-utilizing strains, L. lactis subsp. lactis biovar diacetylactis and Leuconostoc spp. [7]. Industrial starters are mainly based on a single species, namely Lactococcus lactis for most cheeses [42,43]. Another genus involved in starter cultures is represented by the genus Bifidobacterium [44]. Bifidobacteria are Grampositive bacteria that show a high G+C content, over 50%, comprehending fermentative microorganisms producing several acids including lactate [22].
Non-starter bacteria cultures are secondary addition to the cheese matrix and influence the aging phase, with interactions with starter bacterial cultures still present. The whole microbiota, composed by starter and non-starter microorganisms, develops the flavor and texture of cheese. The whole microbial population of the secondary addition differs from the starter culture, being a combination of prokaryotic and eukaryotic microorganisms represented by bacteria, yeasts and molds. The choice of the secondary addition depends on the type of cheese to be produced [45]. Nonstarter lactic acid bacteria are composed by mesophilic lactobacilli and pediococci. They represent important components of the microbial communities of most cheeses during ripening and are characterized by a scarce adaptability to grow in milk and to contribute to acidification processes during cheese formation [46]. Considering their metabolism, cheeses host the facultatively heterofermentative group of non-starter lactic acid bacteria lactobacilli, also defined facultatively heterofermentative lactobacilli, with the most diffused Lactobacillus casei, Lb. paracasei, Lb. plantarum, Lb. rhamnosus and Lb. curvatus. Concerning the genus Pediococcus, Pe. acidilactici and Pe. pentosaceus are the most common species which can be found in cheese [45,47]. Nonstarter lactic acid bacteria include strains essential for producing the characteristic flavours of traditional cheeses. Thermophilic lactic acid bacteria, with an optimal growth between 30 °C and 45 °C are best known for their role in yoghurt-type products and as ripening agents in Swiss-type and Italian cheeses [7]. The numbers of non-starter lactic acid bacteria are reported to be higher in Cheddar cheeses made from raw milk than in those from pasteurised milk [7]. Intense flavor are present in raw milk cheeses, suggesting that the indigenous non-starter lactic acid bacteria play an important role in flavour development. Moreover, they have been shown to contribute to the formation of small peptides and amino acids, which are the precursors for the flavour components. During ripening Cheddar cheese, and for most cheeses, up to 20 strains form the non-starter lactic acid bacteria population [45]. Complex dynamics are present among non-starter lactic acid bacteria, with one non-starter lactic acid bacteria strain that can affect the development of another. The outgrowth of a Lb. rhamnosus strain, added on purpose as an adjunct in a Cheddar cheese trial, could be retarded by a simultaneously added Lb. casei strain. The development of a secondary flora depends also on the properties and composition of the starter used. The presence of a Leuconostoc species in the starter appears to affect the development of adventitious non-starter lactic acid bacteria [7]. Adjunct cultures are microorganisms added to cheese for purposes other than acid formation. Selected adjunct non-starter lactic acid bacteria cultures can accelerate ripening, producing desirable flavour. Adjunct cultures may eliminate defects inhibiting growth adventitious nonstarter dangerous lactic acid bacteria. In Cheddar cheese, cheese flavour was improved by adding strains isolated from raw milk cheese and able to increase formation of amino acids. Cheese made from milk inoculated with strains of Lb. plantarum or Lb. casei subsp. pseudoplantarum received the best gradings [48]. Table 1 [49-54] refers to starter and non-starter microbial cultures involved in cheese and other dairy products fermentation and successive ripening and flavor development
Table 1: Starter and non-starter cultures of microorganisms that play a role in dairy products.
|
Microorganisms |
Dairy Products |
Functions/Features |
Time of starter Addition |
References |
|
Lactic Acid Bacteria |
||||
|
Lactobacillus acidophilus |
fermented milk, acidophilus milk |
acidification and flavor development |
primary addition |
[49] |
|
Lactobacillus brevis |
kefir |
acidification and flavor development |
primary addition |
[50] |
|
Lactobacillus casei |
artisanal cheese, fermented milk |
flavor development |
secondary addition |
[51,52] |
|
Lactobacillus casei ssp. casei |
probiotic fermented milk |
acidification and flavor development |
primary addition |
[49] |
|
Lactobacillus delbrueckii ssp. bulgaricus |
yoghurt, fermented milk |
acidification and flavor development |
primary addition |
[49] |
|
Lactobacillus delbrueckii ssp. lactis |
Italian and Swiss cheese types |
acidification and flavor development |
primary addition |
[51,52] |
|
Lactobacillus helveticus |
semi‐hard, hard cheese |
flavor development and health benefits |
secondary addition |
[49] |
|
Lactobacillus johnsonii |
fermented milk |
acidification and flavor development |
primary addition |
[49] |
|
Lactobacillus kefir |
kefir |
acidification and flavor development |
primary addition |
[50] |
|
Lactobacillus kefiranofacies |
kefir |
acidification and flavor development |
primary addition |
[50] |
|
Lactobacillus paracasei |
artisanal cheese |
flavor development |
secondary addition |
[51,52] |
|
Lactobacillus paracasei spp. paracasei |
kefir |
acidification and flavor development |
primary addition |
[50] |
|
Lactobacillus plantarum |
artisanal cheese, kefir |
flavor development |
secondary addition |
[51,52] |
|
Lactobacillus rhamnosus |
fermented milk |
acidification and flavor development |
primary addition |
[49] |
|
Lactococcus lactis ssp. cremoris |
most cheeses |
acidification and flavor development |
primary addition |
[51,52] |
|
Lactococcus lactis ssp. lactis |
most cheeses, butter, buttermilk |
acidification and flavor development |
primary addition |
[51,52] |
|
Lactococcus lactis ssp. lactis var. diacetylactis |
cheese, butter, buttermilk |
diacetyl production |
primary addition |
[49] |
|
Leuconostoc lactis |
soft and semi-hard cheese |
flavor development and CO2 production |
secondary addition |
[51,52] |
|
Leuconostoc mesenteroides ssp. cremoris |
soft and semi-hard cheese, butter, buttermilk |
flavor development and CO2 production |
secondary addition |
[49] |
|
Streptococcus thermophilus |
Italian and Swiss cheese types, yoghurt, fermented milk |
acidification and flavor development |
primary addition |
[51,52] |
|
Bifidobacteria |
||||
|
Bifidobacterium bifidum |
yoghurt, fermented beverages, cottage cheeses, ice cream, desserts, cheese |
probiotic activity |
primary addition |
[53,54] |
|
Bifidobacterium longum |
yoghurt, fermented beverages, cottage cheeses, ice cream, desserts, cheese |
probiotic activity |
primary addition |
[53,54] |
|
Propionibacteria |
||||
|
Propionibacterium freudenreichii |
Swiss‐type cheese |
hole formation, flavor development |
secondary ripening |
51,52] |
|
Other Bacteria |
||||
|
Brevibacterium linens |
smear‐ripened cheese |
color, flavor development |
secondary ripening |
[51,52] |
|
Corynebacterium casei |
smear‐ripened cheese |
flavor development |
secondary ripening |
[51,52] |
|
Fungi |
||||
|
Debaryomyces hansenii |
smear‐ripened |
aspect, texture, and flavor development |
secondary ripening |
[51,52] |
|
Geotrichum candidum |
smear‐ripened |
aspect, texture, and flavor development |
secondary ripening |
[51,52] |
|
Penicillium camemberti |
white moldy cheese |
aspect, texture, and flavor development |
secondary ripening |
[51,52] |
|
Penicillium roqueforti |
blue‐veined cheese |
aspect, texture, and flavor development |
secondary ripening |
[51,52]
|
Quality Features and Flavor Development in Cheese
Sensorial properties confer unique characteristics to cheese and depend on the type of milk used, the type of feed supplied to the animal that providing the milk, the practices adopted in the processing of the milk, the ripening environment, the type and duration of ripening, the quantity of microorganisms involved and the role they play in the forming product [4]. The active agents at the base of cheese flavour development are enzymatic reactions of microbial origin that create a balance between different components [55]. The characteristics of the flavour profile of ripened cheeses are mainly affected by proteolysis of caseins and in some types also by lipolysis. The typical cheese flavour results from further degradation of amino acids, due to the pathways for conversion of amino acids by starter bacteria [56]. That is true in cheeses produced from pasteurised milk and using starter cultures under aseptic conditions. However, cheese ripening is influenced by different factors, including the microflora of the raw milk, coagulant, starter cultures and by adventitious contamination of the cheese by non-starter bacteria [42]. Based on sensory evaluation and chemical analysis of cheeses, various groups of volatile compounds have been identified as being responsible for the final taste and aroma of cheese. These compounds comprise fatty acids, esters, aldehydes, alcohols, ketones, sulphur compounds and various other components [57-60]. The starter lactic acid bacteria, Lactococcus and Streptococcus, were the dominant microorganisms during the fermentation period, but as the cheese entered the aging period, the relative abundance shifted with detection of non-starter lactic acid bacteria, which were unidentified members of the family Lactobacillaceae [61]. Starter lactic acid bacteria communities appeared to be significantly high (108 CFU g-1) in the initial state of aging, but the population decreased by two or more logs with further aging (Settanni and Moschetti 2010). On the other hand, nonstarter lactic acid bacteria exhibited continuous growth in the population with further aging [62]. During cheese manufacturing, protein degradation occurs during the aging period and contributes to the flavor, texture, and appearance of the final product [63]. The enzymes of non-starter lactic bacteria contribute to the proteolysis process, represented by the hydrolysis of proteins to small peptides and free amino acids, which are the precursors of flavor-forming reactions [63]. In artisanal cheeses, during the aging period, the non-starter lactic acid bacteria Leuconostoc mesenteroides, Lactobacillus helveticus, Lactobacillus zeae, and Enterococcus spp. were detected in low numbers. These nonstarter lactic acid bacteria contributed to the fermentation of residual lactose or other sugars and were involved in citrate, peptide, and amino acid production, including formation of aromatic compounds, as well as contributing to the aging process [64]. Lactobacillus plantarum was detected during the cheese aging phase and influenced the sensory improvemet characteristics [65]. Non-starter lactic acid bacteria mostly originate from raw milk [66]. The non-starter lactic acid bacteria also have been isolated from various locations within cheese production facilities including floors, drains, and equipment surfaces [67]. Cross contamination can be another source of non-starter lactic acid bacteria because some non-starter lactic acid bacteria found in their cheese samples were found in starter lactic acid bacteria of the other types of cheese that were not part of the study but had been produced in the same facility [45,61].
The yeast Geotrichum candidum, formerly Oidium lactis, represents a key microorganism in the catabolism of triglycerides and casein and also in cheese organoleptic properties and appearance development. G. candidum relates to characteristics associated with carbohydrate, lipid, and amino acid metabolism [68]. First classified as mould, G. candidum has been classified as yeast by Barnett and coll. [69]. The yeast G. candidum is an important component of the microflora of soft cheeses such as Camembert and semi-fresh goat’s and ewe’s milk cheese. G. candidum starts to grow on the surface of the rind of cheese at the beginning of the cheese ripening process, where it contributes to the development of typical cheese flavours. Many enzymes contribute to the action of G. candidum in cheese ripening, with peptidases playing a major role in the breakdown of bitterness and the production of flavour compounds [68]. G. candidum is present in raw milk cheeses and it has been detected in cheese regardless of the type of milk used, that can be indifferently represented by milk from cows, ewes and goats. The most likely source of G. candidum in pasteurised milk cheeses is the environment of the cheese factory, comprehending air, floors and/or walls, equipment, and workers. Commercial strains of G. candidum are available for use as starter cultures for cheese ripening and may be added to the milk, to the brine, or sprayed on the cheese surface [68]. G. candidum, as in the French cheese St. Marcellin, can grow forming a white and uniform surface on the surface at temperatures ranging from 5 to 38 °C and at pH 5.0-5.5. G. candidum lipases are characterized by a high specificity in the use of unsaturated fatty acids as substrates. Concerning proteinases and peptidases of G. candidum, intracellular and extracellular proteolytic activities were highlighted. G. candidum contributes to an aroma of mild cheeses such as Brie and traditional Camembert. In addition, G. candidum can contribute to a decrease in diacetyl production, the main flavour compound in cheese, by means of its diacetyl-reductase activity. G. candidum reduces bitterness through the activity of its aminopeptidases by hydrolysing low molecular weight hydrophobic peptides originating from the degradation of casein by Penicillium camemberti, conferring to the cheese organoleptic properties and appearance that are characteristic of traditional Camembert. G. candidum plays an important role in the competition with undesirable microorganisms in mould fermented cheese [68]. The yeast Debaromyces hansenii is involved in the metabolism of other amino acids [70]. Yeast D. hansenii metabolism evidences a role in the deamination of amino acids present in cheese, to the corresponding keto acids and ammonia (NH3 ), with a consequent increase in pH values [47,71]. The different strains of the entire microbial community of cheese are capable of producing enzymes responsible for the degradation of milk. Furthermore, these enzymes may also be responsible for the production of the aromatic compounds in cheese, thus improving its quality and variety of flavor. The microorganisms of cheese therefore represent crucial elements in the development of aromatic compounds that originate during the ripening and maturing phases [72,73].
Pathogenic Microorganisms in Cheese and Dairy Products
Pathogenic bacteria including Listeria monocytogenes, Escherichia coli O157:H7, Salmonella spp., Staphylococcus aureus, Campylobacter spp. can contaminate raw milk, persist in this environment and be transmitted through cheese and dairy products, threatening human health [74]. Other bacterial pathogens such as Brucella sp. [75] and Clostridium botulinum [76] were detected in dairy products, highlighting the sensitivity of these food products to possible contamination by dangerous microorganisms. In Table 2, examples of pathogenic bacteria contaminating dairy products have been reported [77- 83].
Table 2: Examples of outbreaks caused by bacterial pathogens in fermented dairy products. VTEC: Vero-cytotoxin producing E. coli.
|
Pathogens |
Fermented Dairy Products |
Outbreaks/Causes |
References |
|
Brucella sp. |
Pecorino cheese |
Brucellosis, one of the most important zoonoses in the Mediterranean and Middle East regions /Originated from raw milk and insufficiently ageing |
[75] |
|
Clostridium botulinum |
Yoghurt |
Botulism/Insufficient process of conserve used as a flavour |
[76] |
|
Escherichia coli O157:H7 |
Gouda cheese |
Infection is a major public health concern in North America, Europe, and other areas of the world/Raw milk used to make cheese; handling problems including insufficient ageing |
[77] |
|
Escherichia coli (VTEC) O157:H7 |
Yoghurt |
Possible improperly cleaned pump |
[78] |
|
Listeria monocytogenes |
Hard cheese |
Listeriosis/Postmanufacture contamination |
[79] |
|
Listeria monocytogenes |
Queso fresco |
Listeriosis |
[80] |
|
Salmonella Enteritidis |
Pecorino cheese “primo sale” |
Salmonellosis |
[81] |
|
Salmonella sp. |
Hard cheese |
Salmonellosis/Cheese made from raw milk |
[82] |
|
Staphylococcus aureus |
Sheep milk cheese |
Foodborne outbreaks caused by staphylococcal toxins/Raw milk used in production |
[83]
|
L. monocytogenes can be life-threatening, with outbreaks associated with high mortality rates, typically 20-30% [84]. L. monocytogenes is an opportunistic pathogen that has the ability to survive in wide variety of environments including low temperature (4 °C), high salt, and low pH [85]. Based on the resistance to stress conditions, the L. monocytogenes strain could survive in the cheese and is a significant safety concern. The DNA high-throughput sequencing techniques are extremely important to ensure safety of cheese, with the important results that can be achieved rapidly. An important outbreak by L. monocytogenes was registered in southern California in 1985, due to the ingestion of Mexican-style cheese, with pregnant women dying, some along with their unborn babies, and mothers losing their newborn infants [86,87]. An important listeriosis outbreak associated with Hispanic-style cheese occurred in 2013 [88]. Application of wholegenome sequencing revealed that cheese was the vehicle of infection and that a bacterial strain isolated from the surrounding environment, showed a high similarity with the outbreak isolate [45,88]. Between October 2002 and February 2003, two outbreaks caused by the strain Escherichia coli O157:H7 and associated with unpasteurized Gouda cheese from a dairy farm in Edmonton, Canada, infected children causing the hemolytic uremic syndrome. Further investigation revealed that the cheese was still contaminated with E. coli O157:H7 104 days after production [89]. In spring 2020, an outbreak of Salmonella Enteritidis occurred in the Marche region (Central Italy), due to the pecorino cheese “primo sale”, produced with raw sheep milk. Microbiological investigation detected Salmonella Enteritidis in animal faeces and in environmental samples, as the raw-milk bulk tanks and in milk taken from single animals [81].
In 2014, an outbreak of Staphylococcus aureus occurred at a boarding school in Switzerland by consuming a soft cheese, Tomme, produced from raw cow milk. The outbreak caused symptoms of abdominal pain, violent vomiting, severe diarrhea, and fever [90]. Campylobacter jejuni is capable of contaminating raw milk and a member of this species has difficulties during isolation due to demanding growth requirements [91]. Outbreaks due to Campylobacter spp. can be detected in the consumption of raw milk (EFSA, 2018). In Germany, in 2018, a high percentage of all reported foodassociated outbreaks originated from bacterial strains Campylobacter spp. and most of them resulted from consumption of raw milk. Strains Campylobacter spp. can survive unfavorable environmental conditions by shifting in the viable but non-cultivable state (VBNC), remaining viable and maintain their infectious potential [92]. Real-time PCR (qPCR) offered a highly sensitive culture-independent quantification method, allowing the determination of the highest possible residual risk present, while the minimal risk is indicated via plate count cultures, by colony-forming unit counts [93].
Defenses against Pathogenic Microorganisms
Aging for 60 days is believed to improve the antimicrobial properties of lactic acid bacteria. Lower levels of pathogenic bacteria, including L. monocytogenes, E. coli O157: H7, Salmonella spp. and Campylobacter spp. were found in cheese samples tested, supporting the hypothesis that 60-day maturation of raw milk cheese can improve microbiologically safe cheeses [94]. The Hazard Analysis Critical Control Point (HACCP) plan in the dairy sector have significantly contributed to the reduction of the incidence of foodborne illnesses linked to the consumption of cheese [95]. However, dairy products of unsatisfactory or borderline quality, according to European Community (EC) recommendations for the presence of pathogens Salmonella spp., S. aureus, E. coli and L. monocytogenes, can still be detected. Eukaryotic microorganisms such as yeasts and molds can be common contaminants of cheeses and dairy products. In particular, these microorganisms can pose a serious health hazard due to their ability to produce mycotoxins [6]. Control of microbiological safety of cheese can be achieved by the action of microorganisms with antimicrobial capabilities and protective effects in cheese [6]. Milk and dairy products colonization by natural microbial communities also helps to inhibit growth of pathogenic bacteria. Lactic acid bacteria are the most significant microorganisms of raw milk cheese and can inhibit the growth of pathogens [96]. Variation of the microbial composition of organic acid-producing bacteria in raw and pasteurized milk influence the survival of L. monocytogenes. In pasteurized milk, bacteria producing organic acid are more common and growth of L. monocytogenes is slower [97]. The contribution of lactic acid bacteria on inhibiting the growth of foodborne pathogens was highlighted in presence of E. coli that evidenced a decrease to less than 1% after the addition of commercial starter cultures [45,61]. Bacteriocins are peptides with antimicrobial activity produced by a wide diversity of bacteria. Those produced by Gram-positive bacteria are classified into class I, containing heavily modified (lanthionine-containing) peptides called lantibiotics, and class II, containing non-modified peptides or peptides with minor modifications, the latter consisting in disulfide bond formation or circularization [98]. Bacteriocins can be rapidly degraded by proteases in the gastrointestinal tract and thus they have no effects to human gut microbiota [99].
Nisin is a class I bacteriocin produced by lactococci and represents the best characterized bacteriocin from lactic acid bacteria. Nisin, listed as E234, can be added as a food additive and is the only bacteriocin with a regulatory approval from the US Food and Drug Administration and European Union. Nisin presents a broad activity spectrum against different pathogenic bacteria, including Listeria sp., enterococci, staphylococci, streptococci, Clostridium sp., C. jejuni, Helicobacter pylori and antibiotic-resistant strains of Neisseria gonorrhoeae [100]. Stability of nisin depends on pH and can be subjected to degradation by proteolytic enzymes in cheese, as was observed in Cheddar cheese after 6 months of ripening, with a significant depletion of antimicrobial activity. Lacticin 3147 is a bacteriocin with two-component broad-spectrum antimicrobial peptide produced by Lactococcus lactis subsp. lactis DPC3147 evidencing a high stability over a wide range of pH. No decrease in lacticin antimicrobial activity was detected in Cheddar cheese over 6 months of ripening. Nevertheless, lacticin 3147 was not effective for controlling L. monocytogenes contamination on surfaces of smear-ripened cheese [101]. Pediocin AcH represents a broad-spectrum anti-listerial bacteriocin that belongs to class II, produced by Pediococcus acidilactici present in many cheeses, although with a low efficiency at pH higher than 5.0 [102]. Thermophilin, produced by some strains of Streptococcus thermophilus, is a class II bacteriocin with inhibitory activity against strains belonging to species of the genera Streptococcus, Enterococcus, Lactococcus, Bacillus and Listeria [103]. Macedocin is a bacteriocin produced by Streptococcus macedonicus ACA-DC 198 isolated from Kasseri, a Greek hard-cooked cheese, and belongs to the lantibiotic bacteriocins (Class I). Macedocin inhibits growth of bacterial strains of lactic acid bacteria and of the species Clostridium tyrobutyricum. C. tyrobutyricum strains can produce butyric and acetic acid, CO2 and H2 from fermentation of lactate, causing large defects in semihard and hard cheeses, such as Swiss-type or Gouda, damaging flavor and inducing late blowing of the cheese [104]. Enterocin 4 is produced by Enterococcus faecalis strain INIA 4 during the manufacture and ripening of the Spanish Manchego cheese and is able to inhibits growth of L. monocytogenes strains. This bacteriocin belongs to the heat-stable, non-lantibiotic class II [105]. The presence of bacteriocinogenic strains as “protective cultures” in cheese and dairy products can represent an interesting approach to improve safety of cheese and dairy products and to protect human health. The absence of acquired antibiotic resistance genes with transfer potential in these cultures must also be respected [6].
Probiotics
Probiotics are live microorganisms which confer a health benefit on the host when administered in adequate amounts. The most diffused probiotics are mainly microorganisms from species of the genera Lactobacillus and Bifidobacterium. Probiotics are defined by the FAO/WHO [106] as “live microorganisms which when administered in adequate amounts confer a health benefit on the host”. The beneficial effects in treatment and prevention of various diseases or gut disorders such as inflammatory bowel disease or lactose intolerance are greatly debated in the literature [76]. Bifidobacteria and Lactobacillus acidophilus are most used in functional dairy foods containing probiotics, especially milk, yoghurt, ice cream and desserts [107]. The viability of probiotics is a key parameter for their effectiveness, with the scientific acceptance that the minimum intake to provide a therapeutic effect requires at least 108 -109 CFU g-1 viable cells. Most cheeses seem to be suitable carriers for probiotic bacteria. It is worth mentioning that Lactobacillus sp. probiotics can inhibit the adhesion of L. monocytogenes to the human epithelium, thus avoiding subsequent infections [108].
Conclusion
In this mini-review the microorganisms involved in the development of cheeses and dairy products have been described, focusing on the key role that microorganisms play in the production of safe products and in maintaining their typicality. The most important microbial groups have been reported, from lactic acid bacteria involved in the fermentation and ripening of milk, to yeasts and molds active in imparting flavor during cheese ripening. A description of the pathogenic bacteria has also been reported, providing information on possible probiotics that produce antimicrobial compounds. Insights for future research in the contexts of dairy microbiology may include increased product diversification and in this context, the isolation of new microbial strains from the raw milk environment and the surrounding environment, followed by identification and characterization of isolates, could provide important bases for innovations and to improve the typicality of cheeses and dairy products. Other innovations have emerged, consisting of isolating lactic acid bacteria that produce new bacteriocins to be characterized and used against pathogens, possibly by bio-augmentation processes of lactic bacteria. Furthermore, metagenomic approaches represent recent methods which include DNA extraction from cheese and dairy products; amplification of microbial genes from extracted DNA and subsequent sequencing; followed by the comparison of the sequences and the elaboration of the whole picture of the microbiota. This powerful tool will allow to monitor microbial pathways during milk transformation processes and to record possible contaminating microorganisms, including pathogens. Both the classic microbiological approach based on the culture of microorganisms and the latest metagenomic-based methodologies should be combined during the microbiological study of cheeses and dairy products, in order to acquire biotechnological opportunities and obtain a comprehensive view of microbiological characteristics. The combination of cultural and metagenomic approaches can offer important opportunities to improve the characteristics of cheeses and dairy products, while respecting traditions.
References
- Ferrocino I, Rantsiou K, Cocolin L (2022) Investigating dairy microbiome: An opportunity to ensure quality, safety and typicity. Current Opinion in Biotechnology 73: 164-170.
- Codex Standard 283‐1978 (2013) Codex General Standard for Cheese; Adopted in 1973; Revision 1999; Amendments 2006; Codex Alimentarius Commission: Rome, Italy.
- Kosikowski FV, Mistry VV (1997) Cheese and fermented milk foods. (3rd edn), Kosikowski, F V, Edn; LLC: Westport, CT, USA.
- Fox PF, Guinee TP, Cogan TM, McSweeney PLH (2017) Principal families of cheese. In: Fox PF, Guinee TP, Cogan TM, McSweeney PLH (Eds), Fundamentals of cheese science. Springer: Boston, MA, USA, pp. 27-69.
- Mayo B, Rodríguez J, Vázquez L, Flórez AB (2021) Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Foods 10(3): 602.
- Grattepanche F, Miescher-Schwenninger S, Meile L, Lacroix C (2008) Recent developments in cheese cultures with protective and probiotic functionalities. Dairy Science & Technology 88: 421-444.
- Wouters JTM, Ayad EHE, Hugenholtz J, Smit G (2002) Microbes from raw milk for fermented dairy products. International Dairy Journal 12(2-3): 91-109.
- Boor KJ, Wiedmann M, Murphy S, Alcaine S (2017) A 100‐year review: Microbiology and safety of milk handling. Journal of Dairy Science 100(12): 9933-9951.
- Larpin‐Laborde S, Imran M, Bonaïti C, Bora N, Gelsomino R, et al. (2011) Surface microbial consortia from Livarot, a French smear‐ripened cheese. Canadian Journal of Microbiology 57(8): 651-660.
- Yunita D, Dodd CER (2018) Microbial community dynamics of a blue‐veined raw milk cheese from the United Kingdom. Journal of Dairy Science 101(6): 4923-4935.
- Feligini M, Panelli S, Buffoni JN, Bonacina C, Andrighetto C, et al. (2012) Identification of microbiota present on the surface of Taleggio cheese using PCR‐DGGE and RAPD‐PCR. Journal of Food Science 77(11): M609-M615.
- Ritschard JS, Amato L, Kumar Y, Müller B, Meile L, et al. (2018) The role of the surface smear microbiome in the development of defective smear on surface‐ripened red‐smear cheese. AIMS Microbiology 4(4): 622-641.
- Irlinger F, Layec S, Hélinck S, Dugat‐Bony E (2015) Cheese rind microbial communities: Diversity, composition and origin. FEMS Microbiology Letters 362(2): 1-11.
- Ceugniez A, Drider D, Jacques P, Coucheney F (2015) Yeast diversity in a traditional French cheese “Tomme d’orchies” reveals infrequent and frequent species with associated benefits. Food Microbiology 52: 177-184.
- Ceugniez A, Taminiau B, Coucheney F, Jacques P, Delcenserie V, et al. (2017) Fungal diversity of “Tomme d’Orchies” cheese during the ripening process as revealed by a metagenomic study. International Journal of Food Microbiology 258: 89-93.
- Lavoie K, Touchette M, St‐Gelais D, Labrie S (2012) Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Science & Technology 92(5): 455-468.
- Ropars J, Cruaud C, Lacoste S, Dupont J (2012) A taxonomic and ecological overview of cheese fungi. International Journal of Food Microbiology 155(3): 199-210.
- Bodinaku I, Shaffer J, Connors AB, Steenwyk JL, Biango‐Daniels MN, et al. (2019) Rapid phenotypic and metabolomic domestication of wild Penicillium molds on cheese. mBio 10: e02445‐19.
- Cavanagh, D, Fitzgerald GF, McAuliffe O (2015) From field to fermentation: The origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiology 47: 45-61.
- Laroute V, Tormo H, Couderc C, Mercier‐Bonin M, Le Bourgeois P, et al. (2017) From genome to phenotype: An integrative approach to evaluate the biodiversity of Lactococcus lactis. Microorganisms 5(2): 27.
- Douglas GL, Klaenhammer TR (2010) Genomic evolution of domesticated microorganisms. Annual Review of Food Science and Technology 1: 397-414.
- Vaughan EE, Heilig HGHJ, Ben-Amor K, de Vos WM (2005) Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiology Reviews 29(3): 477-490.
- Stiles ME, Holzapfel WH (1997) Lactic acid bacteria of foods and their current taxonomy. International Journal of Food Microbiology 36(1): 1-29.
- Fernández M, Hudson JA, Korpela R, de los Reyes-Gavilán CG (2015) Impact on human health of microorganisms present in fermented dairy products: An overview. Hindawi Publishing Corporation BioMed Research International.
- Zhu Y, Zhang Y, Li Y (2009) Understanding the industrial application potential of lactic acid bacteria through genomics. Applied Microbiology and Biotechnology 83(4): 597-610.
- Tamime AY, Robinson RK (1999) Yoghurt Science and Technology. (2 Eds.), Cambridge: Woodhead Publishing Ltd, UK.
- Bintsis T (2018) Lactic acid bacteria: their applications in foods. Journal of Bacteriology & Mycology: Open Access 6(2): 89-94.
- EFSA (2008) Scientific opinion of the panel on biological hazards on the request from EFSA on the maintenance of the list of QPS microorganisms intentionally added to food or feed. EFSA Journal 928: 1-48.
- Wedajo B (2015) Lactic acid bacteria: benefits, selection criteria and probiotic potential in fermented food. Journal of Probiotics & Health 3.
- Law BA (1999) Cheese ripening and cheese flavour technology. In: Law BA (Ed.), Technology of Cheesemaking, Sheffield: Sheffield Academic Press Ltd, England, pp. 163-192.
- Smit G, Smit BA, Engels WJ (2005) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiology Reviews 29(3): 591-610.
- Souza MJ, Ardo Y, McSweeney PLH (2001) Advances in the study of proteolysis in cheese. International Dairy J 11(4-7): 327-345.
- Chopin A (1993) Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiology Reviews 12(1-3): 21-38.
- Curtin AC, McSweeney PLH (2004) Catabolism of amino acids in cheese during ripening. In: Fox PF, McSweeney PLH, Cogan TM, et al. (Eds.), Cheese: Chemistry, Physics and Microbiology, (4th edn), London: Elsevier Academic Press, US, pp. 435-454.
- Fox PF, Wallace JM (1997) Formation of flavour compounds in cheese. Advances in Applied Microbiology 45: 17-85.
- Khalid NM, Marth EM (1990) Lactobacilli-their enzymes and role in ripening and spoilage of cheese: A review. Journal of Dairy Science 73(10): 2669-2684.
- Christensen JE, Dudley EG, Pederson JA, et al. (1999) Peptidases and amino acid catabolism in lactic acid bacteria. Antonie van Leeuwenhoek 76(1-4): 217-246.
- Bintsis T, Robinson RK (2004) A study of the effects of adjunct cultures on the aroma compounds of Feta-type cheese. Food Chemistry 88(3): 435-441.
- Lister J (1878) On the lactic fermentation and its bearing to pathology. Transactions of the Pathological Society of London 29: 425-467.
- Hayek SA, Ibrahim SA (2013) Current limitations and challenges with lactic acid bacteria: A review. Food Science & Nutrition 4(11): 73-87.
- Fox PF, Wallace JM (1997) Formation of flavor compounds in cheese. Advances in Food Microbiology 45: 17-85.
- Fox PF, O’Connor TP, McSweeney PLH, Guinee TP, O’Brien NM (1996) Cheese: Physical, biochemical and nutritional aspects. Advances in Food and Nutrition Research 39: 163-305.
- McSweeney PLH, Fox PF (1997) Chemical method for the characterization of proteolysis in cheese during ripening. Lait 77: 41.
- Miyake T, Watanabe K, Watanabe T, Oyaizu H (1998) Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiology and Immunology 42(10): 661-667.
- Nam JH, Cho YS, Rackerby B, Goddik L, Park SH (2021) Shifts of microbiota during cheese production: impact on production and quality. Applied Microbiology and Biotechnology 105(6): 2307-2318.
- Cogan TM, Barbosa M, Beuvier E, Bianchi-Salvadori B, Cocconcelli PS, et al. (1997) Characterisation of the lactic acid bacteria in artisanal dairy products. Journal of Dairy Research 64(3): 409-421.
- Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM (2001) Recent advances in cheese microbiology. International Dairy Journal 11(4-7): 259-274.
- Lynch CM, McSweeney PLH, Fox PF, Cogan TM, Drinan FD (1996) Manufacture of Cheddar cheese with and without adjunct lactobacilli under controlled microbiological conditions. International Dairy Journal 6(8-9): 851-867.
- Tamime AY (2002) Microbiology of starter cultures. In: Robinson RK (Ed), Dairy microbiology handbook, (3rd edn.), John Wiley & Sons Inc, New York, US, pp. 261-366.
- Kıvanç M, Yapıcı E (2015) Kefir as a probiotic dairy beverage: determination lactic acid bacteria and yeast. International Journal of Food Engineering 1(1): 55-60.
- Parente E, Cogan TM (2004) Starter cultures: General aspects. In: Cheese: Chemistry, Physics and Microbiology, (3rd edn.), Fox P O, Ed.; Elsevier: Oxford, UK, pp. 123-147.
- Fox PF, Guinee TP, Cogan TM, McSweeney PLH (2017) Starter cultures. In: Fox PF, Guinee TP, Cogan TM, McSweeney PLH (Eds.), Fundamentals of Cheese Science. Springer: Boston, MA, USA, pp. 121-183.
- Boylston TD, Vinderola CG, Ghoddusi HB, Reinheimer JA (2004) Incorporation of bifidobacteria into cheeses: challenges and rewards. International Dairy Journal 14(5): 375-387.
- Prasanna PHP, Grandison AS, Charalampopoulos D (2014) Bifidobacteria in milk products: an overview of physiological and biochemical properties, exopolysaccharide production, selection criteria of milk products and health benefits. Food Research International 55: 247-262.
- Delahunty CM, Piggott JR (1995) Current methods to evaluate contribution and interactions of components to flavour of solid foods using hard cheese as an example. International Journal of Food Science and Technology 30(5): 555-570.
- Broome MC, Limsowtin GKY (1998) Starter peptidase activity in maturing cheese. Australian Journal of Dairy Technology 53(2): 79-82.
- Bosset JO, Gauch G (1993) Comparison of the volatile flavour compounds of six European ‘AOC’ cheeses using a new dynamic headspace GC-MS method. International Dairy Journal 3: 423-460.
- Engels WJM, Dekker R, De Jong C, Neeter R, Visser S (1997) A comparative study of volatile compounds in water-soluble fraction of various types of ripened cheese. International Dairy Journal 7(4): 255-263.
- Urbach G (1995) Contribution of lactic acid bacteria to flavour compound formation in dairy products. International Dairy Journal 5(8): 877-903.
- Maarse H, Visscher CA (1989) Volatile compounds in foods: Qualitative and quantitative data. Zeist, TNO-CIVO, Food Analysis Institute, The Netherlands, p. 49.
- Choi J, Lee SI, Rackerby B, Frojen R, Goddik L, et al. (2020) Assessment of overall microbial community shift during Cheddar cheese production from raw milk to aging. Applied Microbiology and Biotechnology 104: 6249-6260.
- Settanni L, Moschetti G (2010) Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiology 27(6): 691-697.
- Wallace JM, Fox PF (1997) Effect of adding free amino acids to cheddar cheese curd on proteolysis, flavour and texture development. International Dairy Journal 7(2-3): 157-167.
- Biolcati F, Ferrocino I, Bottero MT, Dalmasso A (2020) Short communication: high-throughput sequencing approach to investigate Italian artisanal cheese production. Journal of Dairy Science 103(11): 10015-10021.
- Aldrete-Tapia A, Escobar-Ramírez MC, Tamplin ML, Hernandez-Iturriaga M (2014) High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiology 44: 136-141.
- Montel MC, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, et al. (2014) Traditional cheeses: rich and diverse microbiota with associated benefits. International Journal of Food Microbiology 177: 136-154.
- Bokulich NA, Mills DA (2013) Facility specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Applied and Environmental Microbiology 79(17): 5214-5223.
- Boutrou R, Guéguen M (2005) Interests in Geotrichum candidum for cheese technology. International Journal of Food Microbiology 102(1): 1-20.
- Barnett JA, Payne RW, Yarrow D (1983) Yeasts: Characteristics and Identification. (1st edn.), Cambridge University Press, UK.
- Monnet C, Dugat-Bony E, Swennen D, Beckerich JM, Irlinger F, et al. (2016) Investigation of the activity of the microorganisms in a reblochon-style cheese by metatranscriptomic analysis. Frontiers in Microbiology 7: 536.
- El Sheikha AF, Montet D (2014) African fermented foods: historical roots and real benefits. In: Ray RC, Montet D (Eds.), Microorganisms and fermentation of traditional foods. Science Publishers Inc, CRC Press, USA, pp. 248-282.
- Irlinger F, Mounier J (2009) Microbial interactions in cheese: implications for cheese quality and safety. Current Opinion in Biotechnology 20(2): 142-148.
- Zheng X, Shi X, Wang B (2021) A review on the general cheese processing technology. Flavor biochemical pathways and the influence of yeasts in cheese. Frontiers in Microbiology 12: 703284.
- Bastam MM, Jalili M, Pakzad I, Maleki A, Ghafourian S, et al. (2021) Pathogenic bacteria in cheese, raw and pasteurised milk. Veterinary Medicine and Science 7(6): 2445-2449.
- Galbraith NS, Ross MS, et al. (1969) Outbreak of brucella melitensis type 2 infection in London. British Medical Journal 1: 612-614.
- O'Mahony M, Mitchell’ E, Gilbert RJ, Begg NT, Rodhouse JC, et al. (1990) An outbreak of foodborne botulism associated with contaminated hazelnut yoghurt. Epidemiology and Infection 104: 389-395.
- McCollum JT, Williams NJ, Beam W, et al. (2012) Multistate outbreak of escherichia coli O157:H7 infections associated with in-store sampling of an aged raw-milk gouda cheese, 2010. Journal of Food Protection 75(10): 1759-1765.
- Sargeant JM, Hafer DJ, Gillespie JR, Oberst RD, Flood SJ (1999) Prevalence of Escherichia coli O157:H7 in white-tailed deer sharing rangeland with cattle. Journal of the American Veterinary Medical Association 215: 792-794.
- Yde M, Naranjo M, MattheusW, Stragier P, Pochet B, et al. (2012) Usefulness of the European epidemic intelligence information system in the management of an outbreak of listeriosis, Belgium, 2011. Eurosurveillance 17(38): 20279.
- Palacios A, Otto M, Flaherty E, Boyle MM, Malec L, et al. (2022) Multistate Outbreak of Listeria monocytogenes Infections Linked to Fresh, Soft Hispanic-Style Cheese — United States, 2021. Morbidity and Mortality Weekly Report 71(21): 709-712.
- Napoleoni M, Villa L, Barco L, Busani L, Cibin V, et al. (2021) A strong evidence outbreak of salmonella enteritidis in central italy linked to the consumption of contaminated raw sheep milk cheese. Microorganisms 9(12): 2464.
- van Duynhoven YTHP, Isken LD, Borgen K, Soethoudt K, Haitsma O, et al. (2009) A prolonged outbreak of Salmonella Typhimurium infection related to an uncommon vehicle: hard cheese made from raw milk. Epidemiology & Infection 137(11): 1548-1557.
- Bone FJ, Bogie D, Morgan-Jones SC (1989) Staphylococcal food poisoning from sheep milk cheese. Epidemiology and Infection 103(3): 449-458.
- Lecuit M (2007) Human listeriosis and animal models. Microbes and Infection 9(10): 1216-1225.
- Guerreiro DN, Arcari T, O’Byrne CP (2020) The σB-mediated general stress response of listeria monocytogenes: life and death decision making in a pathogen. Frontiers in Microbiology 11: 1505.
- Centers for Disease Control (CDC) (1985) Listeriosis outbreak associated with Mexican-style cheese--California. Morbidity and Mortality Weekly Report 34(24): 357-359.
- Ibarra-Sánchez LA, Van Tassell ML, Miller MJ (2017) Invited review: Hispanic-style cheeses and their association with Listeria monocytogenes. Journal of Dairy Science 100(4): 2421-2432.
- Chen Y, Luo Y, Carleton H, Timme R, Melka D, et al. (2017) Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes isolates associated with an outbreak linked to cheese, United States, 2013. Applied and Environmental Microbiology 83(15): e00633-e00617.
- Honish L, Predy G, Hislop N, Chui L, Kowalewska-Grochowska K, et al. (2005) An outbreak of E. coli O157:H7 hemorrhagic colitis associated with unpasteurized Gouda cheese. Canadian Journal of Public Health 96(3): 182-184.
- Johler S, Weder D, Bridy C, Huguenin MC, Robert L, et al. (2015) Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. Journal of Dairy Science 98(5): 2944-2948.
- Davis KR, Dunn AC, Burnett C, et al. (2016) Campylobacter jejuni infections associated with raw milk consumption — Utah, 2014. Morbidity and Mortality Weekly Report 65: 12.
- Baffone W, Casaroli A, Citterio B, Pierfelici L, Campana R, et al. (2006) Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. International Journal of Food Microbiology 107(1): 83-91.
- Wulsten IF, Galeev A, Stingl K (2020) Underestimated survival of campylobacter in raw milk highlighted by viability real-time PCR and growth recovery. Frontiers in Microbiology 11: 1107.
- Brooks JC, Martinez B, Stratton J, Bianchini A, Krokstrom R, et al. (2012) Survey of raw milk cheeses for microbiological quality and prevalence of foodborne pathogens. Food Microbiology 31(2): 154-158.
- Goulet V, De Valk H, Pierre O, Stainer F, Rocourt J, et al. (2001) Effect of prevention measures on incidence of human listeriosis, France, 1987-1997. Emerging Infectious Diseases 7(6): 983-989.
- Yoon Y, Lee S, Choi KH (2016) Microbial benefits and risks of raw milk cheese. Food Control 63: 201-215.
- Lee J, Seo Y, Ha J, Kim S, Choi Y, et al. (2020) Influence of milk microbiota on Listeria monocytogenes survival during cheese ripening. Food Science & Nutrition 8(9): 5071-5076.
- Umu ÖCO, Bäuerl C, Oostindjer M, Pope PB, Hernández PE, et al. (2016) The Potential of class II bacteriocins to modify gut microbiota to improve host health. PLoS ONE 11(10): e0164036.
- Bernbom N, Licht TR, Brogren CH, Jelle B, Johansen AH, et al. (2006). Effects of Lactococcus lactis on composition of intestinal microbiota: role of nisin. Applied and Environmental Microbiology 72(1): 239-244.
- Morency P, Dubois MJ, Gresenguet G, Frost E, Masse B, et al. (2001) Aetiology of urethral discharge in Bangui, Central African Republic. Sexually Transmitted Infections 77: 125-129.
- O’Sullivan L, O’Connor EB, Ross RP, Hill C (2006) Evaluation of live-culture-producing lacticin 3147 as a treatment for the control of Listeria monocytogenes on the surface of smear-ripened cheese. Journal of Applied Microbiology 100(1): 135-143.
- Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H (2006) The continuing story of class IIa bacteriocins. Microbiology and Molecular Biology Reviews 70(2): 564-582.
- Fontaine L, Hols P (2008) The inhibitory spectrum of thermophilin 9 from streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpGSt, a thiol-disulfide oxidase. Applied and Environmental Microbiology 74(4): 1102-1110.
- Bachmann HP (1999) Factors influencing the germination and growth of Clostridium tyrobutyricum in hard cheese made from silage-free milk. Information from the field of food inspection and hygiene 90: 62-72.
- Nuñez M, Rodríguez JL, García E, Gaya P, Medina M (1997) Inhibition of Listeria monocytogenes by enterocin 4 during the manufacture and ripening of Manchego cheese. Journal of Applied Microbiology 83(6): 617-677.
- FAO/WHO (2006) Health and nutrition properties of probiotics in food including powder milk with live lactic acid bacteria.
- Moslehishad M, Ehsani MR, Salami M, Mirdamadi S, Ezzatpanah H, et al. (2013) The comparative assessment of ACE-inhibitory and antioxidant activities of peptide fractions obtained from fermented camel and bovine milk by Lactobacillus rhamnosus PTCC 1637. International Dairy Journal 29(2): 82-87.
- Drolia R, Amalaradjou MAR, Ryan V, Tenguria S, Liu D, et al. (2020) Receptor-targeted engineered probiotics mitigate lethal Listeria infection. Nature Communications 11: 6344.